amep.spatialcor.sfiso#
- amep.spatialcor.sfiso(coords: ndarray, box_boundary: ndarray, N: int, qmax: float = 20.0, twod: bool = True, njobs: int = 1, chunksize: int | None = None, mode: str = 'std', other_coords: ndarray | None = None, accuracy: float = 0.5, num: int = 64, verbose: bool = False) tuple[ndarray, ndarray]#
Calculates the isotropic static structure factor for a given set of coordinates.
Notes
The isotropic static structure factor is defined by
\[S_{3D}(q) = \frac{1}{N}\left\langle\sum_{m,l=1}^N\frac{\sin(qr_{ml})}{qr_{ml}}\right\rangle\]\[S_{2D}(q) = \frac{1}{N}\left\langle\sum_{m,l=1}^N J_0(qr_{ml})\right\rangle\]with \(r_{ml}=|\vec{r}_m-\vec{r}_l|\) and the Bessel function of the first kind \(J_0(x)\).
For mode=’fast’, a rewritten form for homogeneous and isotropic systems is used that reduces the number of computations from \(\mathcal{O}(N^2)\) to \(\mathcal{O}(N)\), where \(N\) is the number of particles [1].
Mode ‘fft’ only works in 2D!!!
References
- Parameters:
coords (np.ndarray of shape (N,3)) – Coordinates.
(3 (np.ndarray of shape) – Boundary of the simulation box in the form of np.array([[xmin, xmax], [ymin, ymax], [zmin, zmax]])
2) – Boundary of the simulation box in the form of np.array([[xmin, xmax], [ymin, ymax], [zmin, zmax]])
N (int) – Total number of particles.
qmax (float, optional) – Maximum wave number to consider. The default is 20.
twod (bool, optional) – If True, the 2D form is used. The default is True.
njobs (int, optional) – Number of jobs for multiprocessing. The default is 1.
chunksize (int, optional) – In ‘std’ mode, the calculation is divided into chunks of this size. The default is 1000.
mode (str, optional) – One of [‘std’, ‘fast’, ‘fft’]. The ‘fft’ mode only works if twod is True. The default is ‘std’.
other_coords (np.ndarray, optional) – Coordinate frame of the other species to which the correlation is calculated. The default is None (uses coords).
accuracy (float, optional) – Accuracy for fft mode. 0.0 means least accuracy, 1.0 best accuracy. The default is 0.5. Note that a higher accuracy needs more memory for the computation. accuracy must be in (0,1].
num (int, optional) – Number of q vectors to average over in ‘fast’ mode. If twod is False, the number of q vectors is equal to num^2. The default is 64.
verbose (bool, optional) – If True, additional information is printed and a progress bar is shown. The default is False.
- Returns:
S (np.ndarray) – Isotropic static structure factor.
q (np.ndarray) – Wave numbers.
Examples
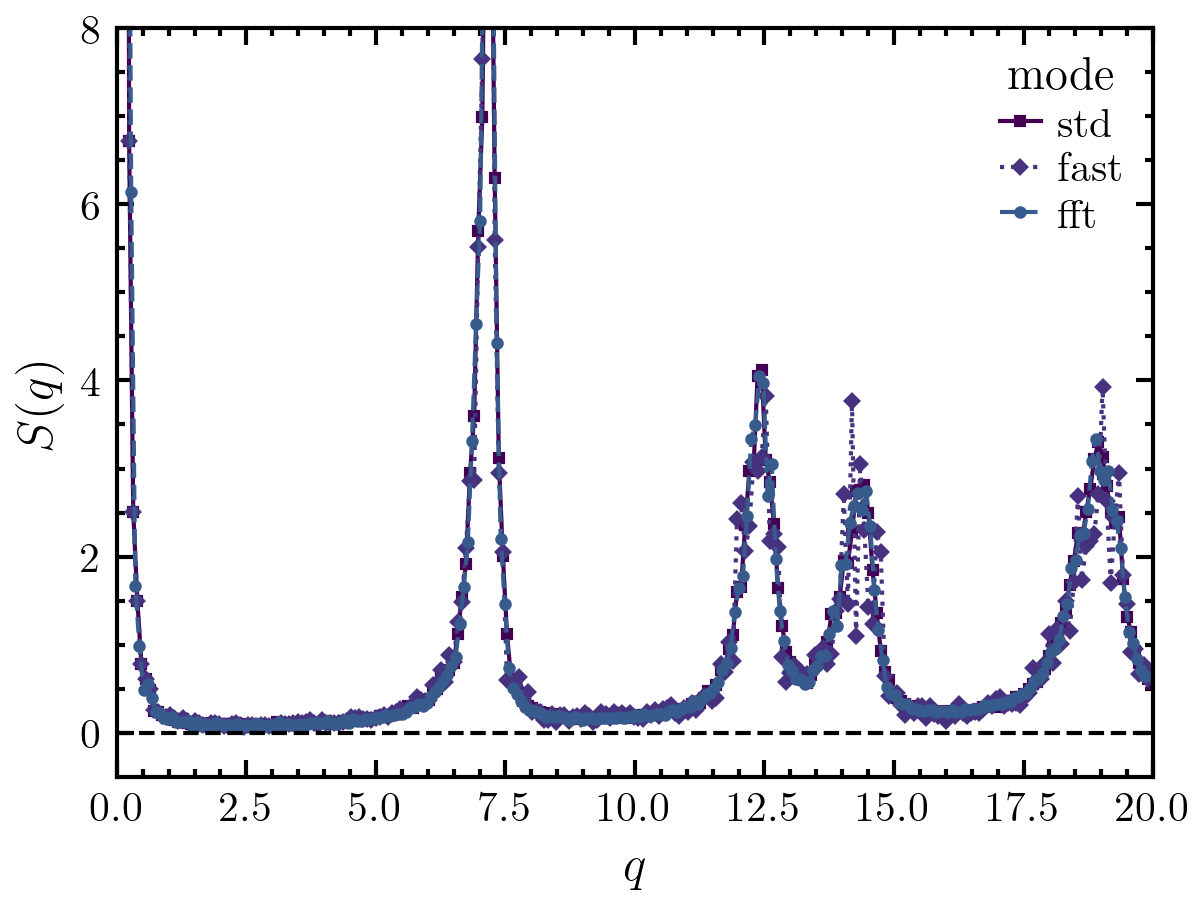
>>> import amep >>> traj = amep.load.traj("../examples/data/lammps.h5amep") >>> frame = traj[-1] >>> S0, q0 = amep.spatialcor.sfiso( ... frame.coords(), frame.box, frame.n(), ... njobs=6, twod=True, chunksize=1000, mode='std' ... ) >>> S1, q1 = amep.spatialcor.sfiso( ... frame.coords(), frame.box, frame.n(), ... njobs=6, twod=True, chunksize=1000, mode='fast' ... ) >>> S2, q2 = amep.spatialcor.sfiso( ... frame.coords(), frame.box, frame.n(), ... njobs=6, twod=True, chunksize=1000, mode='fft', accuracy=0.5 ... ) >>> fig, axs = amep.plot.new() >>> axs.plot(q0, S0, ls='-', label='std') >>> axs.plot(q1, S1, ls=':', label='fast') >>> axs.plot(q2, S2, ls='--', label='fft') >>> axs.set_ylim(-0.5, 8) >>> axs.set_xlim(0, 20) >>> axs.legend(title='mode') >>> axs.set_xlabel(r'$q$') >>> axs.set_ylabel(r'$S(q)$') >>> axs.axhline(0, c='k', ls='--', lw=1) >>> fig.savefig('./figures/spatialcor/spatialcor-sfiso.png') >>>