amep.evaluate.RDF#
- class amep.evaluate.RDF(traj: ParticleTrajectory, skip: float = 0.0, nav: int = 10, ptype: int | None = None, other: int | None = None, **kwargs)#
Bases:
BaseEvaluationRadial pair distribution function.
- __init__(traj: ParticleTrajectory, skip: float = 0.0, nav: int = 10, ptype: int | None = None, other: int | None = None, **kwargs) None#
Calculate the radial pair-distribution function [1].
Works for a one-component system. Takes the time average over several time steps.
Notes
This version only works for one-component systems in 2D. The radial pair-distribution function is defined by (see Refs. [2] [3])
\[g(r) = \frac{1}{\rho N}\sum\limits_{k}\sum\limits_{l\neq k} \left\langle\frac{\delta\left(r-\left|\vec{r}_k(t) -\vec{r}_l(t)\right|\right)}{2\pi r}\right\rangle_t\]References
- Parameters:
traj (ParticleTrajectory) – Trajectory object of particle-based simulation data.
skip (float, optional) – Skip this fraction at the beginning of the trajectory. The default is 0.0.
nav (int, optional) – Maximum number of frames to consider for the time average. The default is 10.
ptype (float, optional) – Particle type. The default is None.
other (float, optional) – Other particle type (to calculate the correlation between different particle types). The default is None.
**kwargs – Other keyword arguments are forwarded to amep.spatialcor.rdf.
Examples
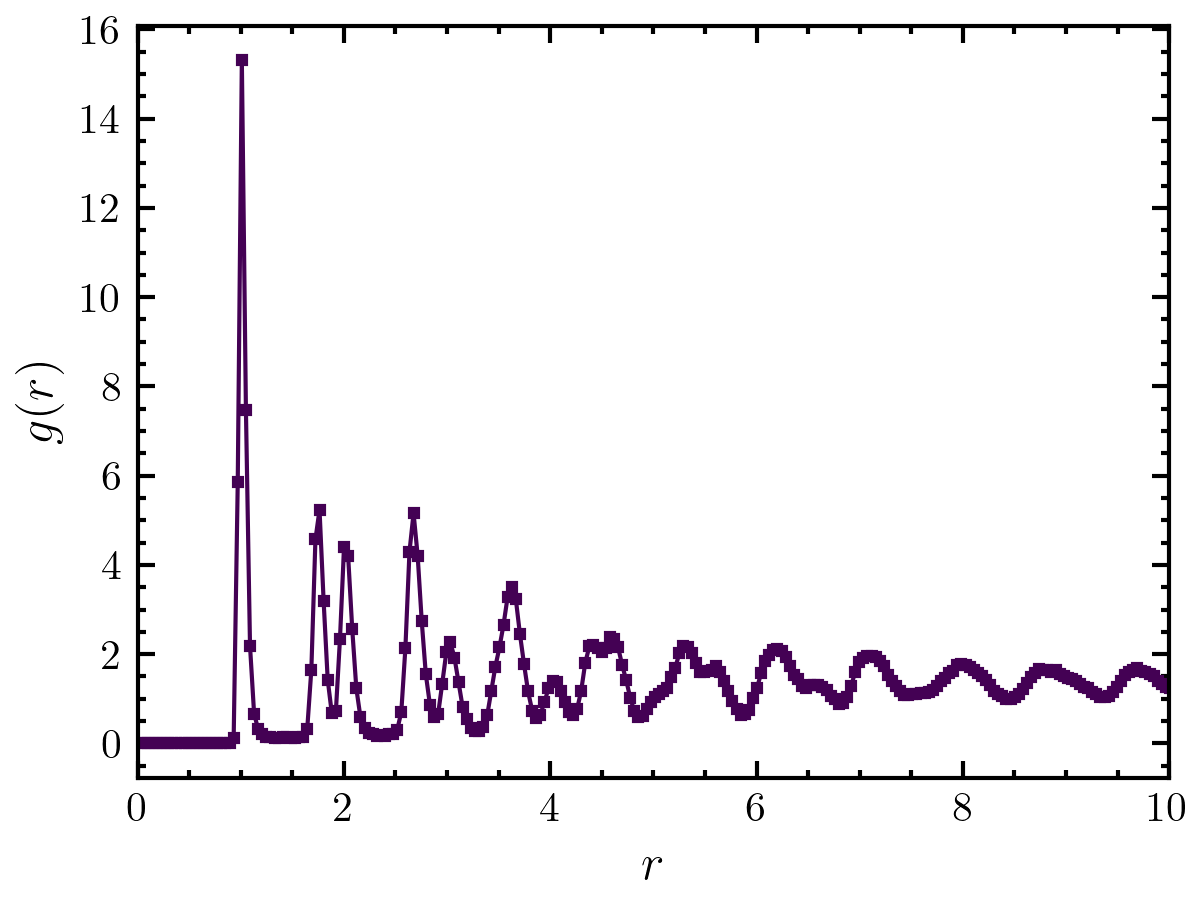
>>> import amep >>> traj = amep.load.traj("../examples/data/lammps.h5amep") >>> rdfcalc = amep.evaluate.RDF( ... traj, nav=2, nbins=1000, skip=0.9, njobs=4 ... ) >>> rdfcalc.save('./eval/rdf.h5') >>> fig, axs = amep.plot.new() >>> axs.plot(rdfcalc.r, rdfcalc.avg) >>> axs.set_xlim(0,10) >>> axs.set_xlabel(r'$r$') >>> axs.set_ylabel(r'$g(r)$') >>> fig.savefig("./figures/evaluate/evaluate-RDF.png")
Methods
__init__(traj[, skip, nav, ptype, other])Calculate the radial pair-distribution function [1].
items()keys()The keys to the evaluation object.
save(path[, backup, database, name])Stores the evaluation result in an HDF5 file.
values()Attributes
Time-averaged RDF (averaged over the given number of frames).
RDF for each frame.
Indices of all frames for which the RDF has been evaluated.
nameDistances.
Times at which the RDF is evaluated.
- property avg#
Time-averaged RDF (averaged over the given number of frames).
- Returns:
Time-averaged RDF.
- Return type:
np.ndarray
- property frames#
RDF for each frame.
- Returns:
Function value for each frame.
- Return type:
np.ndarray
- property indices#
Indices of all frames for which the RDF has been evaluated.
- Returns:
Frame indices.
- Return type:
np.ndarray
- keys() list[str]#
The keys to the evaluation object.
Used so Evaluation-objects can be used as dictionaries.
- property r#
Distances.
- Returns:
Distances.
- Return type:
np.ndarray
- save(path: str, backup: bool = True, database: bool = False, name: str | None = None) None#
Stores the evaluation result in an HDF5 file.
- Parameters:
path (str) – Path of the ‘.h5’ file in which the data should be stored. If only a directory is given, the filename is chosen as self.name. Raises an error if the given directory does not exist or if the file extension is not ‘.h5’.
backup (bool, optional) – If True, an already existing file is backed up and not overwritten. This keyword is ignored if database=True. The default is True.
database (bool, optional) – If True, the results are appended to the given ‘.h5’ file if it already exists. If False, a new file is created and the old is backed up. If False and the given ‘.h5’ file contains multiple evaluation results, an error is raised. In this case, database has to be set to True. The default is False.
name (str or None, optional) – Name under which the data should be stored in the HDF5 file. If None, self.name is used. The default is None.
- Return type:
None.
- property times#
Times at which the RDF is evaluated.
- Returns:
Times at which the RDF is evaluated.
- Return type:
np.ndarray
