amep.cluster.linear_extension#
- amep.cluster.linear_extension(coords: ndarray, box_boundary: ndarray, mass: ndarray, pbc: bool = True, clusters: list | None = None) float#
Calculate the linear extension (end-to-end distance) of one cluster.
Takes the coordinates of each particle in the cluster. Uses the minimum image convention if pbc is True.
Notes:#
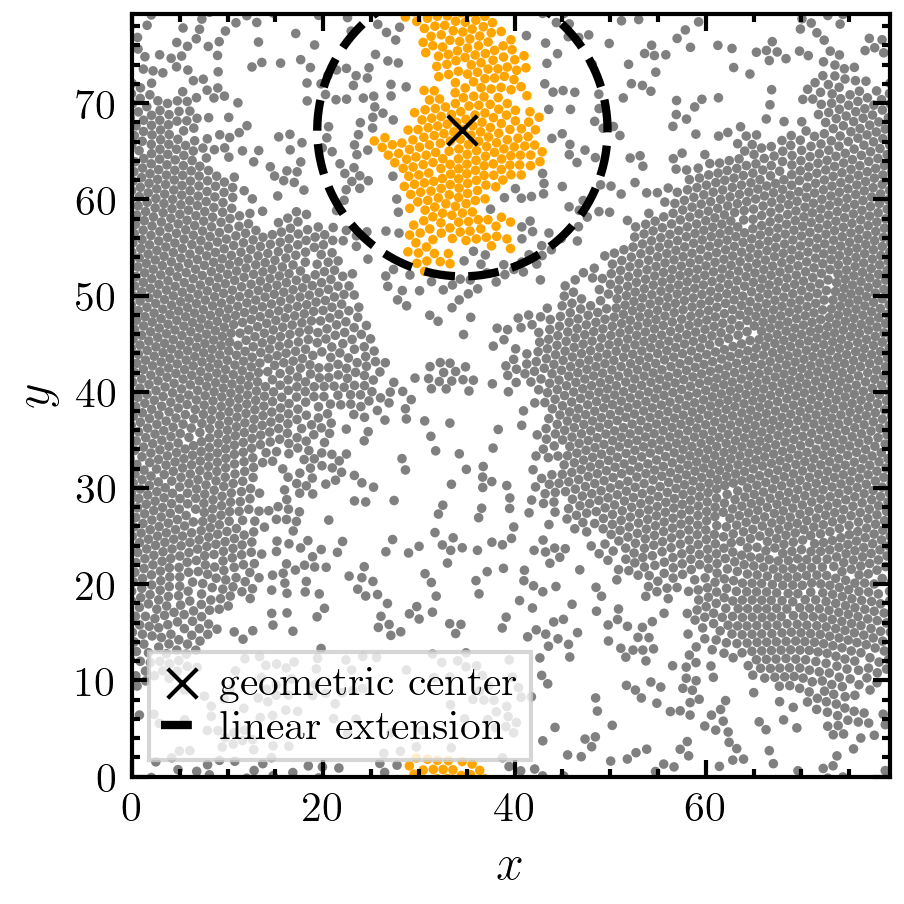
The linear extension is defined as the maximal distance between two particles in the same cluster [1]. This is an O(N²) algorithm. An O(N) implementation (which is implemented in this function) is to calculate twice the in-cluster maximum distance between a cluster particle and the cluster center of mass [2].
References:#
Parameters:#
- coordsnp.ndarray of shape (N,3)
Coordinates of all particles inside the cluster.
- box_boundarynp.ndarray of shape (3,2)
Boundary of the simulation box in the form of np.array([[xmin, xmax], [ymin, ymax], [zmin, zmax]]).
- centernp.ndarray of shape (3,)
Center of the simulation box.
- massnp.ndarray of shape (N,)
Mass of all particles inside the cluster.
- pbcbool, optional
If True, periodic boundary conditions are considered. The default is True.
- clusterslist or None, optional
List of lists, where each list contains the indices of the particles that belong to the same cluster. If None, the geometric center of the given coords is calculated. If not None, the geometric center of each cluster in clusters is calculated, where coords must be the coordinates of the particles in clusters. The default is None.
- returns:
Linear extension of the cluster.
- rtype:
float or np.ndarray of shape (N,)
Examples
>>> import amep >>> import numpy as np >>> traj = amep.load.traj("../examples/data/lammps.h5amep") >>> frame = traj[25] >>> clusters, idx = amep.cluster.identify( ... frame.coords(), frame.box, pbc=True ... ) >>> coords = frame.coords()[idx==1] >>> mass = frame.mass()[idx==1] >>> Le = amep.cluster.linear_extension( ... coords, frame.box, mass, pbc=True ... ) >>> print(Le) 30.3604296244119
>>> les = amep.cluster.linear_extension( ... frame.coords(), frame.box, frame.mass(), ... pbc=True, clusters=clusters ... ) >>> centers = amep.cluster.geometric_center( ... frame.coords(), frame.box, clusters=clusters ... ) >>> i = 1 >>> fig, axs = amep.plot.new(figsize=(3,3)) >>> amep.plot.particles( ... axs, frame.coords()[idx==i], frame.box, color="orange", ... radius=frame.radius()[idx==i] ... ) >>> amep.plot.particles( ... axs, frame.coords()[idx!=i], frame.box, color="gray", ... radius=frame.radius()[idx!=i] ... ) >>> axs.scatter( ... centers[i,0], centers[i,1], color="k", s=50, marker='x', ... label="geometric center" ... ) >>> angle = np.linspace(0, 2*np.pi, 150) >>> x = les[i]/2*np.cos(angle)+centers[i,0] >>> y = les[i]/2*np.sin(angle)+centers[i,1] >>> axs.plot( ... x, y, c="k", ls="--", lw=2, marker="", label="linear extension" ... ) >>> axs.set_xlabel(r"$x$") >>> axs.set_ylabel(r"$y$") >>> axs.legend(frameon=True) >>> fig.savefig("./figures/cluster/cluster-linear_extension.png")